Pipeline to aggregate data from Climatology Lab
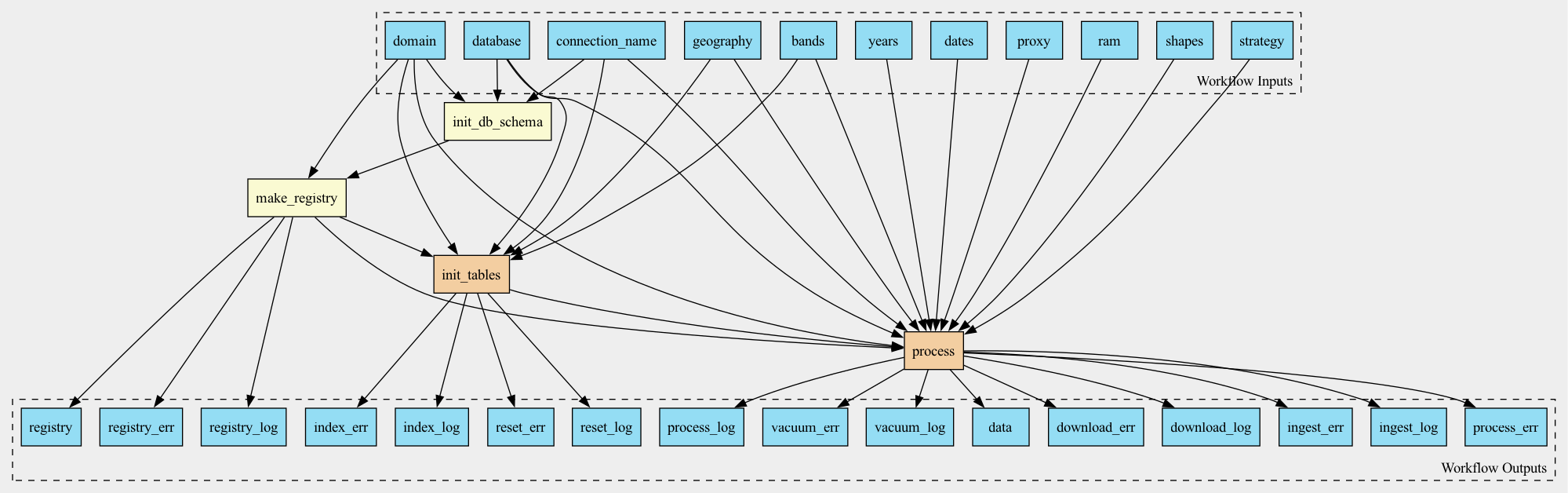
Workflow
Description
This workflow downloads NetCDF datasets from University of Idaho Gridded Surface Meteorological Dataset, aggregates gridded data to daily mean values over chosen geographies and optionally ingests it into the database.
The output of the workflow are gzipped CSV files containing aggregated data.
Optionally, the aggregated data can be ingested into a database specified in the connection parameters:
database.inifile containing connection descriptionsconnection_namea string referring to a section in thedatabase.inifile, identifying specific connection to be used.
The workflow can be invoked either by providing command line options as in the following example:
toil-cwl-runner --retryCount 1 --cleanWorkDir never \
--outdir /scratch/work/exposures/outputs \
--workDir /scratch/work/exposures \
gridmet.cwl \
--database /opt/local/database.ini \
--connection_name dorieh \
--bands rmin rmax \
--strategy auto \
--geography zcta \
--ram 8GB
Or, by providing a YaML file (see example) with similar options:
toil-cwl-runner --retryCount 1 --cleanWorkDir never \
--outdir /scratch/work/exposures/outputs \
--workDir /scratch/work/exposures \
gridmet.cwl test_gridmet_job.yml
Inputs
Name |
Type |
Default |
Description |
|---|---|---|---|
proxy |
string? |
HTTP/HTTPS Proxy if required |
|
shapes |
Directory? |
Do we even need this parameter, as we instead downloading shapes? |
|
geography |
string |
Type of geography: zip codes or counties Valid values: “zip”, “zcta” or “county” |
|
years |
string[] |
|
|
bands |
string[] |
University of Idaho Gridded Surface Meteorological Dataset bands |
|
strategy |
string |
|
Rasterization strategy used for spatial aggregation |
ram |
string |
|
Runtime memory, available to the process. When aggregation strategy is |
database |
File |
Path to database connection file, usually database.ini |
|
connection_name |
string |
The name of the section in the database.ini file |
|
dates |
string? |
dates restriction, for testing purposes only |
|
domain |
string |
|
Outputs
Name |
Type |
Description |
|---|---|---|
registry |
File |
|
registry_log |
File |
|
registry_err |
File |
|
data |
array |
|
download_log |
array |
|
download_err |
array |
|
process_log |
array |
|
process_err |
array |
|
ingest_log |
array |
|
ingest_err |
array |
|
reset_log |
array |
|
reset_err |
array |
|
index_log |
array |
|
index_err |
array |
|
vacuum_log |
array |
|
vacuum_err |
array |
Steps
Name |
Runs |
Description |
|---|---|---|
init_db_schema |
[‘python’, ‘-m’, ‘nsaph.util.psql’] |
We need to do it because of parallel creation of tables |
make_registry |
Writes down YAML file with the database model |
|
init_tables |
creates or recreates database tables, one for each band |
|
process |
Downloads raw data and aggregates it over shapes and time |